A collaboration between Monash University, the University of Technology Sydney, and the University of Queensland has laid the foundations for a new class of antimalarial drugs, with the help of the high-throughput protein crystallography beamline at the Australian Synchrotron.
The work is part of international efforts to defeat the malaria parasite, which infects as many as half a billion people around the world every year and kills more than one million, many of them children.
Dr Sheena McGowan, Dr Corrine Porter and Professor James Whisstock from the ARC Centre of Excellence in Structural and Functional Microbial Genomics at Monash University used the synchrotron to obtain high-resolution crystal structures of a key malarial enzyme in its natural state and the same enzyme bound to potential inhibitors.
The enzyme, a neutral aminopeptidase known as PfA-M1, is involved in the terminal stages of haemoglobin digestion. It is essential for producing the amino acids that the malaria parasite needs in order to grow and develop inside red blood cells. The enzyme is located outside the parasite's main digestive compartment, making it a potentially easier target for drugs.
Prof John Dalton from UTS and Prof Don Gardiner from the University of Queensland showed how a PfA-M1 inhibitor called bestatin and another chemical compound with a similar structure were able to bind to the enzyme's active site, stopping it from functioning and effectively starving the parasite to death.
Their work has revealed features within the enzyme's active site that will now become prime targets for the development of a previously undescribed class of antimalarial therapeutics.
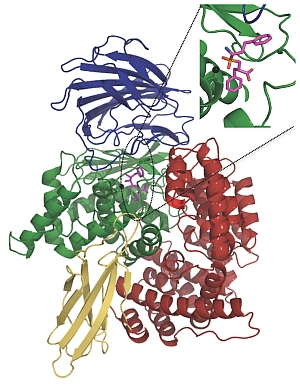 |
A potential new antimalarial drug (shown here in magenta) blocks the active site of a key digestive protein, effectively starving the malaria parasite to death. |
Image courtesy of the ARC Centre of Excellence in Structural and Functional Microbial Genomics at Monash University
